Antibiotics Resistance: An Overview
Rihab FELLAH
Before the antibiotics era, life expectancy was as short as 47 years* due to untimely death including from untreatable infections. In fact, whether you are young or old, a wound as simple as a scratch if infected could mean certain death. However, when these miraculous molecules known as antibiotics came, by chance, to the attention of Mr Fleming and thereafter the rest of the scientific and medical community, everybody thought it was the end of bacterial infections. Little did we know that, these microscopic creatures have the antidote to the venom they are the source of: Resistance to antibiotics, a poetic expression for a great medical problem. If resistance to antibiotics was ever to prevail, will we ever return to that dark era? How could we stop it? Or is it inevitably the consequence of using (not just misusing) these precious molecules?
Introduction
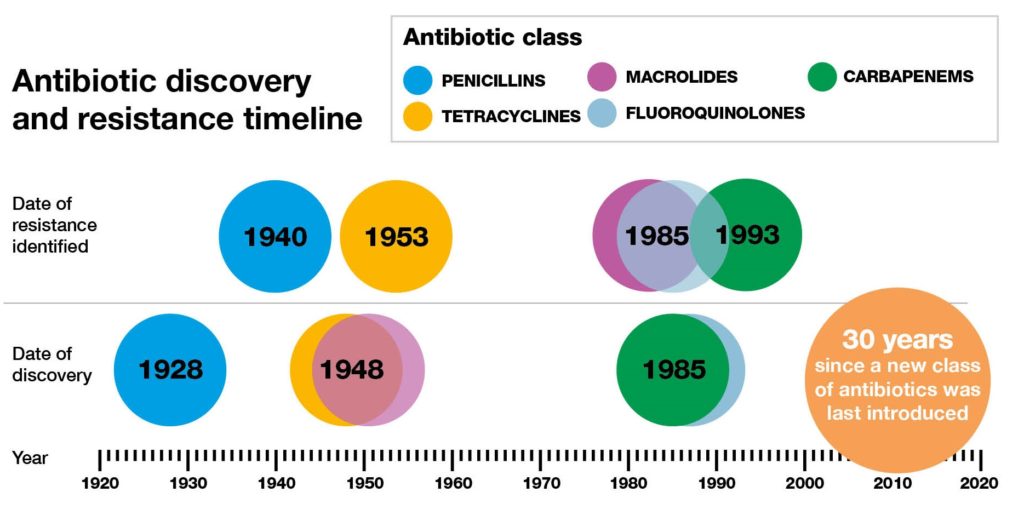
Antibiotics resistance has existed way before the serendipitous discovery of these molecules by Fleming in 19281. Indeed, genes of certain bacteria dating to 30000 years ago that were preserved and isolated in the permafrost of the arctic region. They were found to be holders of resistance genes2. Also, many bacteria that live in aquatic environments subsist on antibiotics, using them as their main source of carbohydrates. It is thought that they may be a potential source of novel resistance genes.2
Antibiotic resistance has been responsible for the reemergence of some diseases in many countries such as the reemergence of multi-drug resistant tuberculosis in the developed world3, the emergence of Multi-Drug Resistant (MDR) typhoid fever in Africa and Asia4-6, and the reemergence of outbreaks of Community acquired Methicillin Resistant Staphylococcus aureus (MRSA) in North America and Australia7.
It is also responsible of concerning infections in hospitals. Nosocomial infections not only are costly but can touch vulnerable individuals such as patients under assisted ventilation in Intensive Care Units (ICU)7.
In Algeria, since the founding of the Algerian Antimicrobial Resistance Network or AARN, the surveillance of Antibiotic resistance throughout the country has permitted detecting the emergence of resistant bacteria such as MRSA, Carbapenem resistant Acinetobacter baumaniii, and Extended spectrum b-lactamase (ESBLs) producing Enterbacteriacæ8,9.
What are antibiotics and how do they work ?
The word antibiotics (ABX) comes from the French word antibiosis meaning “against life”, a term coined in 1889 by the French scientist Jean Paul Vuillemin which means “a substance produced by microorganisms that in small amounts inhibits another microorganism.”10
Technically the synthetic sulfa drugs aren’t antibiotics they are antimicrobial drugs because they are not synthesized by microorganisms1,10.However, in clinical practice we use it to apply to all antimicrobial drugs synthetic or not that inhibit or kill bacteria.
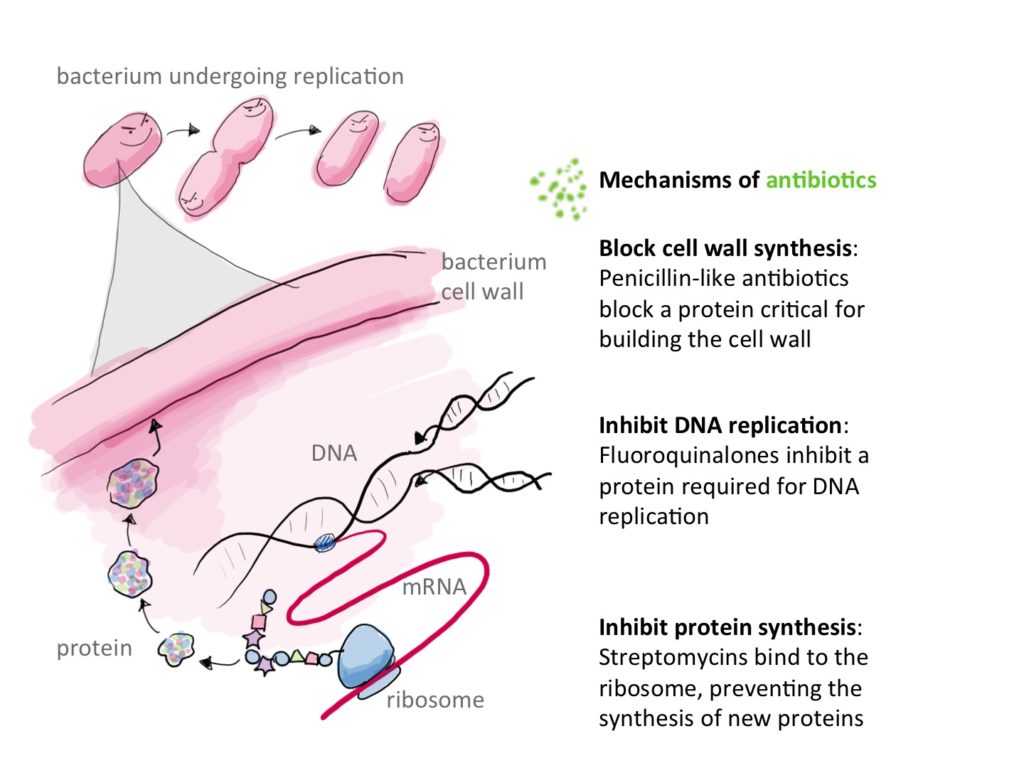
Antibiotics stop or inhibit bacterial growth by targetting many essential functions to these microscopic creatures :
1) By inhibiting cell wall synthesis directly : binding to peptidoglycans and inhibiting their binding to each other. It is the case of Vancomycin and other glycopeptides. Vancomycin binds directly to the D-alanyl-D-alanine of the peptidoglycan halting its synthesis. b-lactams on the other hand, bind to the enzyme responsible for the intertwining of peptidoglycans. This enzyme is appropriately called penicillin-binding-protein12.
2) Inhibiting synthesis of bacterial DNA as is the case of fluoroquinolones which bind to DNA Gyrase, preventing it from participating in DNA metabolism and thus killing bacteria.
3) inhibiting protein synthesis by binding to ribosomal RNA this is the case of many intracellular acting ABX such as those of the group MLSb (Macrolids ,Lincosamids, Streptogramins b), Tetracyclines, Aminoglycosides2,12.
Aminoglycosides’ activity is further enhanced when administered with cell-wall synthesis inhibitors such as glycopeptides (famously the anti-MRSA Vancomycin) and b-lactams molecules. This is what is called a synergistic effect.
4) Inhibiting certain metabolic pathways as is the case of the famous association Sulfamethoxazole and trimethoprim or cotrimoxazole (SMX-TMP) who inhibit folic acid metabolism. Also, the NADPH pathway can be inhibited by metronidazole2,12.
Reproduced from: http://2016.igem.org/wiki/images/2/2d/Antimicrobial_resistance_dundee_igem_2016.jpg, licensed under CC BY
Mechanisms of resistance
Before talking about mechanisms through which resistance is created, transmitted and maintained in the microbiome, we must talk about types of resistance and how bacteria manage to escape the anti-life effect of these molecules.
There exists two types of resistance: high level resistance in which increasing the doses of the antibiotic in question cannot overcome this mechanism. It is seen for example in enterococcus resistance to vancomycin. Enterococcuschanges in its wall of peptidoglycan the site upon which vancomycin acts. Therefore, no matter the concentration of vancomycin present around it, it wouldn’t recognize the changed target and therefore won’t inhibit the synthesis of the bacterial wall, resulting in the survival of resistant enterococci. It’s also the case of resistance mediated by enzymatic inactivation of antibiotics11. Low level resistance, on the other hand, can be overcome by increasing antibiotics concentrations as seen in case of infections by pneumococci having low sensitivity to penicillin. Pneumococcus changes its penicillin binding protein. But it expresses a mosaic of PBPs. Some bind penicillin and some don’t. Therefore, in elevating the concentration of this antibiotic, we can inhibit cell wall synthesis eventually 2,11,12.
It is fascinating to consider how many bacterial populations manage to escape the anti-infective effect of antibiotics. Some of these ingenious mechanisms are summarized in the paragraph that follows. They are in no way exhaustive, but serve only as examples to spike your interest into considering them and their clinical consequences.
Enzymatic inactivation is the mechanism of resistance that is most widespread among bacteria. Not only does it concern most classes of antibiotics it can be combined to other mechanisms and thus responsible for multidrug resistances2.
Resistances to the biggest family of antibiotics: the b-lactam family can be enzymatic. It is both seen in Gram positive and Gram negative bacteria.
Among Gram positive cocci, we have Staphylococcus aureus’s resistance to methicillin (ancient antistaphylococcal b-lactam that is equivalent to oxacillin, no more used in clinical practice but the term is often referred to in antibiotics susceptibility tests) with which it can resist a wide range of b-lactams. This same resistance can be transferred from staphylococcus to enterococcus of the same microbiome.
The b-lactamases are the main resistance mechanism in Gram negative bacilli (GNB). The ESBLs are the most notorious type since their discovery in the late 80s. They’ve been detected in many GNB but are predominant mostly in Klebsiella species, and Escherichia coli13.
When this type of resistance is identified in GNB infections, clinicians feel obliged to use carbapenems as an alternative. Therefore a risk of selecting b-lactamases of the broad spectrum type that can hydrolyse carbapenems ensues. This results in carbapenemase-producing GNB, MDR-bacteria that are classified third in the WHO priority pathogens list for Research & Discovery of new antibiotics13-15.
Enzymatic inactivation of aminoglycosides is quite different from that of b-lactams. Aminoglycosides are de-activated by enzymes who either acetylate, phosphorylate or adenylate them. Therefore they’re not hydrolysed as is the case of b-lactams. They’re structurally altered. Also, some of these enzymes can be bifunctional. This means aminoglycosides can be doubly altered. What a hit! This resistance is often expressed among staphylococci and enterococci2.
The danger lying in these enzymatic inactivations is presented in two case figures. The gene for the resistance coding for b-lactamases is often co-transported with the genes for aminoglycosides’ resistance. This means that enterococci become resistant for two major antibiotics used in treating this species. This knowledge combined with the fact that enterococci are naturally resistant to third generation cephalosporins, puts us clinicians in a tough spot especially if we are facing Vancomycin Resistant Enterococci (VRE). This particular subgenra of this microbial family is often selected in a microbiome by the use of vancomycin on MRSA infections. How, you may ask? Well, in killing Vancomycin sensitive MRSA we end up selecting in the gut microbiota, Vancomycin resistant enterococci. Usually VRE’s growth is inhibited by its competition with Vancomycin sensitive enterococci VSE. When vancomycin is used for long periods of time this selection occurs and the predominant enterococci of the gut become those resistant to vancomycin. In the end this results in what is known in the English language as a dilemma16.
Another antibiotic-hydrolyzing enzyme is erythromycin esterase that confers high level resistance against macrolides. It is often transferred with NMD1 (New Delhi Metallo-b-lactamase) gene2,11. A gene that codes for many antibiotic-inactivating enzymes such as b-lactamases too. Staphylococci that are resistant to macrolides can also be resistant to lincosamides and streptogramins which are macrolide-like molecules. This bacterium uses structure-modifying enzymes and deactivates these antibiotics by adenylation or acetylation2.
Other than enzymatic inactivation, the use of efflux pumps is a different mechanism of resistance. This is seen for resistance against cyclines and quinolones. Rarely is this mechanism isolated. For example, the resistance to quinolones often combines the induction of an efflux mechanism and the change in the antibiotic site of action which in this case is the DNA gyrase. Efflux pumps can be used in resisting the use of blactams2.
Changing the target site is mostly seen in the pneumococcus. This alpha-hemolytic streptococcus which is notoriously known for causing severe pneumonia and meningitis can alter his PBPs by acquiring new genes coding for new ones. Not only does it express these novel molecules but it also continues to express its older PBPs resulting thus in a phenotype consisting of a mosaic of PBPs. It is why this type of resistance is a low level one. In fact, a usual dose of penicillin doesn’t inhibit cell wall synthesis but higher doses can. These new PBPs can also be inducible by the presence of b-lactams in the environment. Therefore a subsequent failure to treatment after an initial favourable response must lead to suspect such a scenario especially if resistent pneumococcus has been identified in you community2.
Altering the target is how enterococci resist vancomycin. Vancomycin work on D-alanyl-D-alanine portion of a peptidoglycans thus halting their synthesis. Enterococcus becomes resistant by changing this target to D-alanyl-D-lactate. The gene transferring this type of resistance is called VanA2.
MRSA expresses, as aforementioned, an oxacillinase via mecA gene transported in the SCCmec cassette that is responsible for many resistances among this genus including resistances to penicillins, streptomycin, erythromycin and tetracyclines. Additionally, It can acquire via horizontal conjugation the VanA gene thus becoming either intermediarily susceptible to vancomycin (VISA) or completely resistant to it (VRSA). This has been described in Japan and the USA but remains a rare encounter due to fitness cost2,7(which we will explain later).
When a combination of these mechanisms is met in a bacterium, high levels of resistance occur. This is often seen in many subtypes of bacteria. In E.coli and in Pseudomonas spp. for instance, which are GNB, a low cell wall permeability is inherently present because of their thick LipoPolySaccharidic (LPS) outer membrane. In Pseudomonas spp, this low permeability is particularly efficacious because the porins of this bacterium have low affinity to ABX and are highly specific and selective of their substrate. Add to these obstacles, a secretion of inactivating enzymes by the bacterium such as Extended spectrum b-lactamases or aminoglycosides’ modifying enzymes and the trick is played. If one mechanism of antibiotic resistance is a low level one, the combination of all these resistance mechanisms will inevitably be responsible for much higher levels of resistance2,17.
Also, if , by some fortune, an antibiotic is capable of escaping a resistance mechanism, and this particular resistance is expressed highly in the whole inoculum of the invading bacteria, its success will be short lived and no matter the concentration used, the battle will be lost eventually. Let’s imagine a penicillin having escaped a staphylococcus’s b-lactamase and having managed to kill that particular microbe. It will only manage to lyse one bacterium in a whole army. The subsequent win will cause the release of all bacterial enzymes including b-lactamases in the extracellular milieu. The result will be doubling the threat for the molecule. It will have to face not only the enzymes of its defeated foe but of its still-living (and still-producing-enzymes) brethren2.
Going back to Pseudomonas , one of the largest-genome-holding bacteria. Its genome is as big as that of a fruit fly. (I remind you that we are still talking about a single cell organism!). MDR Pseudomonas has managed to be second in the WHO priority pathogens list for Research & Discovery of new antibiotics. Why? It can acquire, quite rapidly, resistance genes and manages to keep them. It is true that Pseudomonas is not a common pathogen and is only evoked in certain situations (Cystic Fibrosis, Chronic Obstructive Pulmonary Disease, Catheter-dwelling infections and hospital-acquired and healthcare-associated infections as well as infections of patients in ICUs). It is a pathogen to be reckoned with due to its high pathogenicity and the low arsenal of molecules available to treat it. This pathogen can manage to evade even the few molecules to which it is susceptible by its adaptive capacity to its environment. It can produce a biofilm that protects a group of cells in the bacterial inoculum from antibiotic exposure. This group called persister cells, manages to survive and not only becomes resistant to said antibiotic but also maintains infection and causes relapse17.
Reproduced from: https://i0.wp.com/sitn.hms.harvard.edu/wp-content/uploads/2016/01/Fig1-Mech-of-antibio.jpg licensed under CC BY-SA-NC
How is resistance generated?
Resistance is the natural consequence of using antibiotics. In fact, through point-mutations in bacterial genome, and in a whole population of rapidly and continuously dividing bacteria, some will express variations in the targets upon which ABX act. Once this population is exposed to ABX, the selection of the resistant sub-population is bound to happen. Consequently, an infection by this subpopulation will be automatically resistant to this previously used antibiotic2.
However, point mutations which are termed microevolutionary events remain rare and are not the main source of antibiotic resistance.
The macroevolutionary events are more frequent. These events happen through an exchange of resistance genes via horizontal transfer (between bacteria form the same genus or from different genera) or vertical transfer (from mother to daughter bacteria).
If they are frequent, macroevolutionary events do not occur easily. This is due to the fact that bacteria protect their DNA from foreign genetic material. Evidently. They use restricting enzymes that work as controlling officers of DNA modification. They are strain specific and can degrade any form of genetic information that doesn’t have the proper protective sequences (i.e. the proper passport)2.
Another way this protection happens is by the use of the nobel-worthy-concept that is the CRISPR Cas9 system. CRISPR that stands for Clustered Regularly Spaced Palindromic Repeats, recognizes any nucleic information not in accord with its own code. It transcribes it then into a crRNA that will be translated and integrated into a nuclease specific to the target in order to degrade it. Of interest to our subject, it has been observed that bacteria lacking this protective system are more likely to express multi-drug-resistant genes than others with it.
Even with these obstacles, transfer of R genes (resistance genes) still happens and through many ways.
The most common and most studied way is for sure plasmids. It is a mobile circular DNA that is transferable between bacteria via a phenomenon called conjugation. This DNA’s replication is independent from that of the bacterial DNA2.
The transmission of R genes can also happen via transposons. They ensure a transfer either from chromosome to chromosome in an independent manner as they can ensure such transfer via the use of plasmids, or via the use of bacteriophage DNA2.
Last, but not least are integrons. Unlike plasmids and transposons, they cannot transfer genetic material in an independent manner .They help the insertion of foreign DNA into bacterial chromosome or into the plasmid via their unique integrase function. They can also be present in a transposon. They function as recombination hotspots: sites in bacterial DNA, a transposon, a plasmid or a bacteriophage DNA in which foreign DNA can be inserted2.
If resistance genes are so valuable, why don’t all bacteria express them? Or at least why don’t all bacteria exposed to ABX express them?
Keeping and/or expressing R genes whether in the chromosome or in the form of plasmids isn’t something given to all bacteria. It often comes with a cost. A fitness cost.
It happens that R genes are considered to be a metabolic burden. They cost energy and when they are rarely used (because ABX exposure can be a rare event in a bacterium’s short life) they can be shed away (especially plasmids) because they do not help with survival.
In a colony of bacteria, those with costly R plasmids don’t multiply as fast as Plasmid-free ones because of this metabolic burden. Therefore it is thought that in the absence of ABX exposure, there’s a natural selection of antibiotic-susceptible Plasmid-free Bacteria.
We conclude that the fitness cost (metabolic burden) of having R genes is only worth it when there’s repeated exposure to ABX. That is the case in hospitals and intensive care units.
Nevertheless, some research proclaims that the horizontal transfer of R genes can be faster and therefore can overcome this problem. The conjugation of plasmids and R genes through the microbiome might be faster than the rate of death of Plasmid-holding bacteria2,18.
In knowing how resistance is generated and sustained, we come to have this idea that all microscopic living organisms and bacteria within our body and our environment are directly affected by our individual and sometimes collective use and misuse of these anti-living molecules that are called antibiotics. For in targeting one species or pathogen by our use of ABX, we end up generating resistances in that particular pathogen as well as in different colonizers of our microbiome and that of the people around us.
In understanding how dynamic antibiotic resistance is, we can fully comprehend the necessity to preserve these molecules, discover new ones and look for alternatives in order to solve the problem at hand.
How do we preserve ABX usage?
The answer is simple. We need to promote a better prescription of ABX by following basic principles of antibiotic therapy. These rules are discussed in many references some of which we recommend you to read10,16. They are in no way exhaustive. They change in time with the emergence of new research on how resistances are created, maintained and how they could be reversed.
Alternatives to ABX
I would like to finish this article with a brief mention of alternatives to antibiotics. Though this armamentarium is only available abroad and might never reach our country for their use remains limited even in high-income countries, it is comforting to know, that through scientific thinking there’s always a solution.
Every couple of days bacteriophages kill half the population of bacteria worldwide. Since antibiotics were offered to us by bacteria resisting other bacteria, scientists thought of exploiting another enemy of the bacterial world which is bacteriophage.
It is a virus specific to bacteria and in replicating inside its specific host, it kills it through a lytic cycle. Not only is it specific to a certain type of bacteria, It doesn’t infect human cells which adds to its benefits. As a matter of fact, bacteriophages have long since been part of the human viriome (the part of human microbiome that consists of viruses). They help keep commensal bacteria in order by killing those wanting to take over (kill the winner scenario). They can be used by certain genera of bacteria against others when they are threatened by them (kill the competition scenario). It is rather safe to say that these long lived allies of ours can be of help in our battle against resistant bacteria. They are still under investigation. The use of antimicrobial peptides is also undergoing investigation. Surely, certain peptides used by animals, amphibians and certain bacteriocins used by some types of bacteria are being investigated for their antimicrobial properties and are considered a promising alternative for antibiotics.
Vaccines have always rid us from pathogens that cannot be treated. Since bacteria are becoming more and more like viruses with fewer therapeutics at hand, the need for vaccines is more necessary today than ever. That is why this preventive therapeutic arm remains an important one in the face of the ever-growing threat of resistance1,19.
Conclusion
If nothing, the COVID-19 pandemic has taught us how fragile we are in the face of contagious pathogens that have no therapy. Today, our awareness of antibiotic resistance as a major threat to public health should be greater than ever. Nonetheless, our fear of this viral pathogen has only exacerbated our bad habits of antibiotic misuse. Azithromycin’s misuse during this pandemic (even after evidence proving its inefficacy alone or in association to other molecules has emerged)20 bears witness to that. It is our hope that through this review, dear reader, your awareness of your responsibility as a physician when using this particularly-important therapeutic arm has grown. You can impact the future of patients and the outcome of many bacterial infections in your society, by simply thinking your subscriptions through.
References
*CDC. National Centre for Health Statistics. Life Expectancy. https://www.cdc.gov/nchs/fastats/life-expectancy.htm
1-Tortora G, Funke B, Case C. Microbiology. 12th ed. Pearson Education, Inc; 2016. Chapter 20: Antimicrobial drugs; 549-550.
2-Opal S M , Pop-Vicas A. Molecular Mechanisms of Antibiotic Resistance in Bacteria. In: Bennett J E, Dolin R, Blaser M J. Mandell, Douglas, And Bennett’s Principles And Practice Of Infectious Diseases. 9th ed. Philadelphia, PA: Elsevier; 2019. 222-239.
3-WHO.Global tuberculosis report. 2020. Available from : https://apps.who.int/iris/bitstream/handle/10665/336069/9789240013131-eng.pdf.
Available in play store : https://play.google.com/store/apps/details?id=uk.co.adappt.whotbreport. And app store : https://apps.apple.com/us/app/tb-report/id1483112411.
MDR resistant Typhoid Fever:
4-Yan M, Li X, Liao Q, et al. The emergence and outbreak of multidrug-resistant typhoid fever in China. Emerg Microbes Infect. 2016 Jun 22;5(6):e62.
5-Hendriksen RS, Leekitcharoenphon P, et al. Genomic signature of multidrug-resistant Salmonella enterica serovar typhi isolates related to a massive outbreak in Zambia between 2010 and 2012. J Clin Microbiol. 2015 Jan;53(1):262-72.
6-Kariuki S, Gordon MA, et al. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015;33 Suppl 3(0 3):C21-C29.
7-Lee A, de Lencastre H et al. Methicillin-resistant Staphylococcus aureus. Nature Reviews Disease Primers. 2018;4(1).
Resistance in Algeria:
8- Baba Ahmed-Kazi Tani Z, Arlet G. Actualité de la résistance aux antibiotiques chez les bacilles à Gram négatif en Algérie. Pathologie Biologie. 2014;62(3):169-178.
9- Tali Maamar H. Surveillance antibiotic resistance in Algeria: challenges achievements and prospects. Presentation presented at; 2018; 1st Annual congress of the Algerian Society of Infectiology – 5th congress of Federation Arab Societies of Clinical Microbiology and Infectious diseases – Algiers. Available from : https://www.sai-dz.com/upload/Presentation%20SAI%2027%20octobre%20%202018.pdf.
Recommended reading on principles of antibiotics use and prescriptions:
10- Spellberg B. Principles of Antiinfective Therapy. In : Bennett J E, Dolin R, Blaser M J. Mandell, Douglas, And Bennett’s Principles And Practice Of Infectious Diseases. 9th ed. Philadelphia, PA: Elsevier; 2019. 211-221.
16-Cunha CB. Principles of Antimicrobial Stewardship. In: LaPlante KL, Cunha CB, Morrill HJ, Rice LB, Mylonakis E. Antimicrobial Stewardship:Principles and Practice. London ,UK: CABi; 2017. pp1-7.
11-Levinson W. Review of medical microbiology and immunology. 13th ed. McGraw-Hill Education; 2014. Chapter 11: Antimicrobial Drugs: Resistance; 221-244.
12-CMIT. Généralités sur les antibiotiques.In E.PILLY 27e Edition : ALINÉA Plus Ed; 2020.
13- Tamma PD, Rodriguez-Bano J. The Use of Noncarbapenem b-Lactams for the Treatment of Extended-Spectrum b-Lactamase Infections. Clin Infect Dis. 2017 Apr 1;64(7):972-980.
14-WHO. WHO priority pathogens list for Research & Discovery of new antibiotics. 2017. Available from : https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
15-Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ. 2016;:1-19.
17-Pang Z, Raudonis R, Glick B et al. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnology Advances. 2019;37(1):177-192.
18-Lopatkin AJ, Meredith HR, Srimani JK, Pfeiffer C, Durrett R, You L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat Commun. 2017 Nov 22;8(1):1689.
19-Czaplewski L, Bax R, Clokie M et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis. 2016 Feb;16(2):239-51.
20- Hossain A, Raknuzzaman M, Tokumura M. Coronavirus (COVID-19) Pandemic: Concern about Misuse of Antibiotics. Journal of Biomedical Analytics. 2020;3(2):19-23.